AllPrep DNA/RNA FFPE Kit (50)
カタログ番号 / ID. 80234
特徴
- 最小限のサンプルから最大限の結果を実現
- 品質を損なうことなく DNAとRNA を遊離
- DNA と RNA を効果的に分離
- 同一 FFPE サンプルにおいて DNA/RNAを包括的に解析
製品詳細
パフォーマンス
図参照
原理
AllPrep DNA/RNA FFPE Kit は、FFPE組織切片からのゲノムDNA とトータルRNAの同時精製用に特別にデザインされています。生体サンプルを分割する必要があるほかの精製法とは異なり、本法ではサンプルを分割せずに高純度DNAおよびRNAが得られます。DNAおよびRNAを別々に精製するためにサンプルを分割すると、性質の異なる可能性のある細胞集団からDNAおよびRNAを精製することになります。また、FFPEサンプルは貴重かつ回収が困難で、量が限られているために、同一サンプルからのDNAおよびRNAの精製はサンプルの浪費を回避することができます。
固定と包埋条件のために、通常FFPEサンプル中の核酸は著しく断片化され、新鮮あるいは凍結したサンプルから得られた核酸よりも分子量が低くなることが頻繁にあります。同一のFFPEサンプルからのDNAおよびRNAを分離する際の大きな問題は、断片化DNAが短く、部分的に一本鎖であり、インタクトなDNAよりもRNAに類似していることです。この断片化DNAの特性のためにDNAおよびRNAの物理的な分離は困難です。AllPrep DNA/RNA FFPE Kitは、特許申請中の溶解法により同一FFPEサンプルからDNAとRNAを別々に遊離できます。
またFFPEサンプル中の核酸はホルムアルデヒドにより化学的に修飾されます。ホルムアルデヒドによるクロスリンクはゲル電気泳動やラボオンチップ解析などの通常の品質管理アッセイでは検出されませんが、酵素的な解析を強く阻害します。AllPrep DNA/RNA FFPE Kitは、DNAやRNAを分解することなくホルムアルデヒドによるクロスリンクを切断するように最適化されています。
操作手順
図参照
アプリケーション
AllPrep DNA/RNA FFPE Kitは、DNAやRNAを分解することなくホルムアルデヒドによるクロスリンクを切断するように最適化されています。 しかしFFPEサンプルから精製した核酸は、高分子のDNAや完全長RNAを必要とするダウンストリームアッセイには推奨されません。いくつかのアプリケーションでは断片化した核酸を使用できるように実験を変更する必要があります(例えばPCRやRT-PCRで短い増幅産物が得られるようにプライマーをデザイン)。cDNA合成にはoligo-dTプライマーの代わりに遺伝子特異的なプライマーの使用を推奨しています。遺伝子特異的なプライマーの使用が不可能な場合は、ランダムプライマーを使用します。
裏付けデータと数値
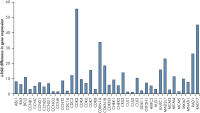
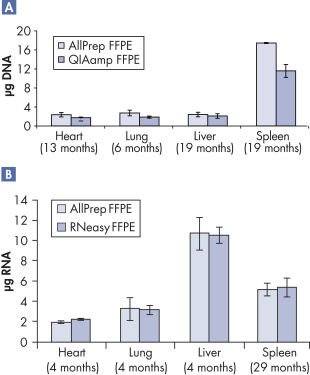
Purification of DNA and RNA from FFPE samples.
仕様
| 特徴 | 仕様 |
|---|---|
| applications | PCR, qPCR, real-time RT-PCR, microarray |
| elutionvolume | RNA: 14-30µl ; DNA: 30-100µl |
| purificationoftotalrnamirnapolyamrnadnaorprotein | RNA (miRNA) and DNA |
| processing | Manual (centrifugation) |
| format | Spin column |
| numberofprepsperrun | 50 |
| sampleamount | max. 4*10 µm sections or 2*20 µm sections |
| technology | Silica technology |
| timeperrunorperprep | 6h for 10 samples, including sectioning |
| yield | Varies |
| mainsampletype | FFPE tissue samples |